3.) Protein Story Phase: Globins
Climbing the Mountain
Globins are a fascinating family of proteins involved in safely carrying gases like oxygen or carbon dioxide throughout the body.
The resources below will introduce the basics of globin structure and function. You will then choose a specific protein story related to globins that you want to model. You will read a scientific research article, find an globin protein structure file, and design a 3D printed physical model of a globin protein.
Note: If you skipped the Training Phase and need to review basic protein structure-function concepts, visit the Training Phase Protein Structure Resources.
Recommended Models for the Globin Protein Story Phase
The MAPS program revolves around using physical models as teaching tools. So if at all possible, we encourage you to use the recommended models below for the globin protein story phase.
All recommended models are available to borrow free of charge through the MSOE Lending Library, or are available for purchase through 3D Molecular Designs. Use the discount code below to receive a 10% MAPS discount on your 3D Molecular Designs purchases.

1. Introduction to Globin Structure and Function
In this section, we will explore the basic structure and functions of globins, including why we need them, how they safely capture gases, and how their amino acid sequence relates to their folded shape.
Globin OverviewThe video below describes how gases like oxygen and carbon dioxide are carried in our blood, to and from our lungs and each cell in our bodies.
Carrying Gases in Our Blood
As the video explains, it is critical that oxygen gets to all the cells in our body so that cellular respiration can convert glucose and oxygen to ATP that is the energy that drives all the important functions of our cells and bodies. In addition, carbon dioxide needs to be carried out of the body so that our blood doesn’t get too acidic which will interfere with these functions as well. Why is this? Because the state of the blood affects the cells and the proteins in them. Proteins are the machines that carry out all the important functions of cells and 1) they often require ATP to drive their function and 2) they need a neutral pH (~7.4) to maintain their specific structure which, as you are learning, is necessary for its function!

Myoglobin - the First Protein Structure
Myoglobin is the first protein to have its 3D structure revealed by the use of X-ray crystallography in the late 1950's. Below are two nice articles that review myoglobin and the discovery of its structure.
\As you might find in your reading on myoglobin, controversy still remains as to the necessity of myoglobin in humans and other mammals, who, unlike whales for example, do not have to spend long periods of time underwater. Diving mammals require a mechanism to store oxygen for long periods of time which is what myoglobin does. But do humans need it? Does it perform an important function during exercise perhaps? This might be a question your team will want to explore for a final project.
All globins contain a Heme Group. This special molecule has a very specific shape that is perfectly suited to holding gas molecules. The video below describes this shape in more detail.
An Introduction to the Heme Group
The Globin Fold Activity
The heme group is held in place by a very important globin structure known as the Globin Fold. It is composed of two alpha helices in a V shape that bracket the heme group on both sides. You learned in the video on hemes that the function of these prosthetic groups are determined largely by the protein environment that surrounds them. We are going to model the globin fold in which the heme is embedded.
This will be a lot more fun with the use of physical models! So if at all possible, we encourage you to use the Betaglobin Folding Kit or the Amino Acid Starter Kit for this section.
- The Betaglobin Folding Kit is available to borrow (free of charge) from the MSOE Lending Library.
- The Betaglobin Folding Kit is available to purchase from the company 3D Molecular Designs.
- The Amino Acid Starter Kit is available to borrow (free of charge) from the MSOE Lending Library.
- The Amino Acid Starter Kit is available to purchase from the company 3D Molecular Designs.
If you have access to a Beta-globin Folding Kit, follow the instructions for folding the 'green segment' of the kit. Then you can proceed to the Globin Fold summary video below.
If you have access to the Amino Acid Starter Kit, use the toober and 2 histidine side chains. Find 2 colors of tape to mark your toober. Your toober represents the middle section of a globin protein and you are going to mark the beginning and end of 3 alpha helices with tape.
- With one color of tape, mark off the beginning and end of 3 alpha helices: 3"-8", 11"-27", 32"-47".
- With a 2nd color of tape, make a mark at 18" and one at 46".
- Make a right-handed helix between each of the helical segments you marked in #1.
- Tug on the ends slightly so that there is some space between the turns of the helices.
- Place the histidines on the marks you made in #2.
- Make a bend in the section between the 2 large helices with the histidines to form a "V" shape and have the histidines face each other.
- Construct a heme group using whatever materials you wish. You can use the templates images below as a guide. To make the heme to scale with the toober globin fold you have constructed, the overall size should be about 2.5" x 3.5".

Take your time with this activities! There is a lot to think about. Don’t be afraid to ask each other questions, as the more you question, the more you learn!
Summary of the Heme Group and the Globin Fold
Myoglobin Sequence Alignment Activity

Start by reading The Stereochemistry of the Protein Myoglobin by H.C. Watson. This describes the analysis that was done using the coordinate data collected from the structure determination experiment in order to better define secondary structures and other conformational details.
Don't let it scare you. It is pretty readable for the most part, and don't get bogged down in the heavy stuff! Focus primarily on Sections 1, 4, and 5.
Once you have read the article, use the links below to guide you through the myoglobin sequence alignment activity. Then watch the summary video to review what you have studied.
Summary of the Myoglobin Sequence Alignment Activity
Globin Sequence Alignment Activity
After analyzing the amino acid sequence of myoglobin and highlighting the secondary structure on the sequence, you are going to compare that with other globins, namely alpha and beta globin, which are the globins in hemoglobin. We have also added neuroglobin for teams interested in this recently discovered globin!
2. Hemoglobin Proteins
In this section we are going to explore one of the most well known globin proteins, hemoglobin. This key protein is responsible for safely transporting molecular oxygen from our lungs, through the blood, to every cell in our body.
The Discovery of Hemoglobin StructureIn the previous section, we explored the structure of myoglobin. You learned that John Kendrew solved the crystal structure of myoglobin. However, he did it by utilizing methods and findings from the experiments that Max Perutz was carrying out to solve the structure of hemoglobin.
In 1962, they shared the Nobel Prize in Chemistry for their work (the same year Watson, Crick, and Wilkins won the Nobel Prize in Physiology or Medicine for the structure of DNA).
You can read about their accomplishment at the Nobel Prize Website
In the video below, Max Perutz describes the research himself, along with a few of his colleagues. If you read the Nobel Prize summary above, you will know the identity of the brash, unnamed graduate student he refers to who dared tell him that his "model was fundamentally flawed"! Hint: he is someone you have probably heard of before...
Max Perutz 1914-2002: "the godfather of molecular biology"
Forty years after Perutz and Kendrew received their Nobel, Kurt Wuthrich received his for using Nuclear Magnetic Resonance (NMR) imaging to solve the structure of hemoglobin and other macromolecules in solution. This adds insight to structures derived from crystals because most proteins are suspended in solution and are able to flex and move slightly so comparisons of crystal structures with NMR structures can explain how proteins move and what parts are involved in their function.
You can learn more about their accomplishment at the Nobel Prize Website, as well as from the video below.
Interview with Kurt Wüthrich
This last video is interesting because of the interest Max Perutz demonstrates in new research, even as his career was coming to an end, and the encouragement he gave to young scientists.
Grabbing Max Perutz' attention: Kurt Wüthrich, 2002 Nobel Laureate in Chemistry
Other Notable Events in the History of Globins
Eighty years ago, Max Perutz began studying hemoglobin. It took him 22 years to solve the structure! Over the next 25 years, many other important discoveries were made that helped us better understand the complex structure-function relationship of hemoglobin. Current hemoglobin research is continually improving and fine-tuning our understanding of the intricacies of this fascinating molecule. There is plenty of research and molecular stories from which you can choose to explore as you Model a Protein Story!
1937 - Perutz begins working on the structure of hemoglobin
1959 - Perutz solves structure of hemoglobin
1962 - Perutz and Kendrew receive Nobel Prize in Chemistry
1965 - Monod/Wyman/Changeaux develop allosteric model of hemoglobin (MWC)
1975 - Fung and Ho identify important salt bridges in deoxyhemoglobin
1985 - Wuthrich develops an NMR method for structural determination of hemoglobin in solution
2002 - Wuthrich receives Nobel Prize in Chemistry
We have studied myoglobin structure-function so that it will be easier to understand the much more complex structure-function relationship of hemoglobin. We know that myoglobin is very similar in structure to alpha and beta globin, so you might be surprised that hemoglobin is so much more complicated. It all stems from the fact that myoglobin is comprised of a single subunit while hemoglobin is made up of 4 subunits, 2 alpha and 2 beta globins. Quaternary structure adds complexity!
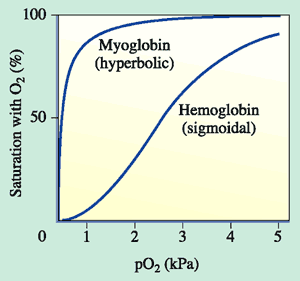
Early on, it was known that hemoglobin was special because of experiments like this one below. If hemoglobin or myoglobin are incubated in solutions with different concentrations (partial pressures, pO2) of oxygen, they fill their hemes with oxygen to varying extents. When all hemes are bound to oxygen, the globins are said to be fully saturated. As you can see from the graph, myoglobin binds oxygen at very low levels of oxygen and becomes fully saturated at about the partial pressure of oxygen at which hemoglobin is only 50% saturated. We will discuss why this is a good thing for our bodies later. You will also notice that the shapes of the curves are different, myoglobin is a hyperbola, and hemoglobin is a sigmoid "s" shape. Max Perutz described this observation in an interesting way: "This strange sigmoid shape signifies that oxygen-free molecules (deoxyhemoglobin) are reluctant to take up the first oxygen molecule but that their appetite for oxygen grows with the eating". Isn't that an interesting metaphor?
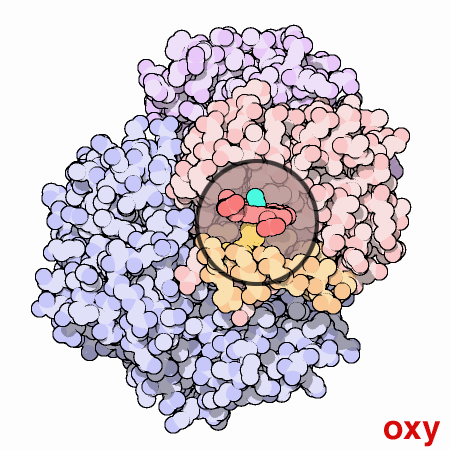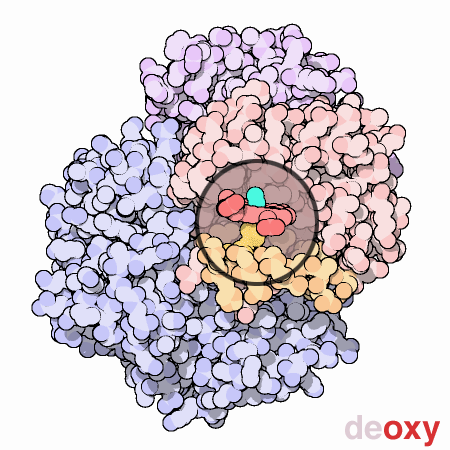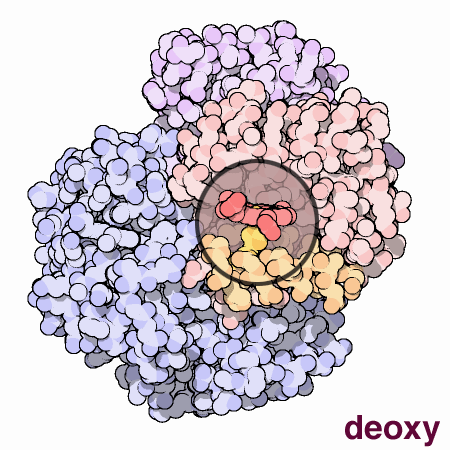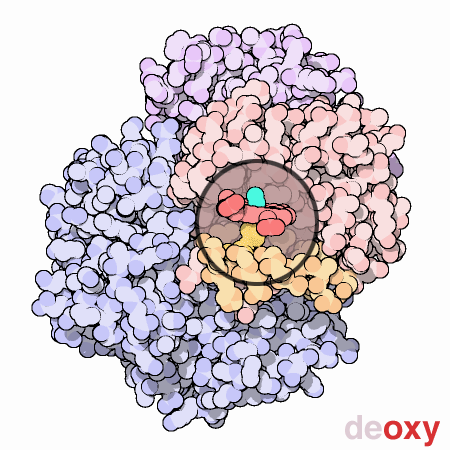
To summarize, each myoglobin molecule can bind one molecule of oxygen and it does that with pretty much the same affinity or "appetite" until it all molecules are bound to oxygen or saturated. Hemoglobin is different. Compare the slope of the hemoglobin curve at pO2 = 1, 2.5, and 5. The slope is lowest at 1 and 5 and highest at 2.5. That means that, as our friend Max stated, hemoglobin is "reluctant" to bind the first oxygen, but once it does, it makes it easier for the 2nd, 3rd, and 4th to bind. This is called cooperativity. Likewise, it is reluctant to give up oxygen (look at the curve from right to left) but once the first molecule of oxygen is released, the others quickly follow.
Interestingly, Jacques Monod, one of the scientists (with Jacob and Lwoff) who received the Nobel Prize in Physiology or Medicine in 1965 for their work on bacterial gene regulation with the lac operon (you may have heard about this in Bio class...), was able to see similarities in the way that hemoglobin bound oxygen to inducible enzymes that he had been studying. He, along with colleagues Wyman and Changeaux, first described the allosteric behavior of hemoglobin in 1965 with their MWC model.
Allostery is basically when something binds to a protein and it either enhances or inhibits binding of another substance to that protein in a different place. You might remember learning about allosteric inhibition in certain enzymes. Proteins that have these sorts of interactions are multimeric, symmetrical proteins that exist in more than one conformation.
Hemoglobin is multimeric (4 subunits), has symmetry, and can exist in 2 different conformations. One conformation is when oxygen is not bound, called deoxyhemoglobin or the T state, while the other is when oxygen is bound, called oxyhemoglobin or the R state. Check out this cool GIF from the Protein Data Bank (below). It gives you an idea of the difference between the deoxy and oxy state. At this point, I just want you to understand that when in the deoxy state, hemoglobin has a low affinity for oxygen or in Max Perutz's words, it has a low "appetite" for oxygen. In the oxy state, it has a high affinity or "appetite" for oxygen. We will discuss more about these 2 states and what affects this transition between them in an upcoming lesson.
Before we spend more time understanding allosteric conformations of hemoglobin, we need to back up and think big picture for a bit. The most important role of hemoglobin is the ability to deliver a sufficient amount of oxygen to metabolically active tissues. Of course, all tissues and cells need oxygen but we will focus on skeletal muscle because, when theses tissues are active, during exercise, they require a large amount of oxygen. How is this achieved? Adaptation to exercise is a very complex process that involves the nervous, respiratory and cardiovascular systems and more, but that is beyond the scope of this program. How is hemoglobin oxygen binding altered depending on tissue needs?
This is a fascinating part of the hemoglobin story. As you remember, myoglobin is an oxygen storage protein found in the muscle and it is also able to transport oxygen within the large muscle cell for ATP generation required for muscle contraction. It binds oxygen with high affinity while hemoglobin binds oxygen with lower affinity. Why is that a good thing? If hemoglobin bound oxygen with the same or greater affinity as myoglobin, it would have a difficult time handing the oxygen off to myoglobin in the cell so that difference in affinity helps deliver oxygen to the muscle cells. Instead, the higher oxygen affinity of myoglobin in effect 'pulls' oxygen off the hemoglobin and onto the myoglobin so that it can get it to the mitochondria of the muscle cell.

Carbon Dioxide and pH
There are allosteric effectors that help promote the release of oxygen to the cells that require it. Remember in the video on Gas Transport we discussed how CO2 is converted to HCO3- and H+ so that the hydrogen ions increase and pH decreases in the venous blood as it removes the waste products from cells making ATP. It turns out that high CO2 and low (acidic) pH shifts the sigmoidal oxygen dissociation curve to the right and decreases the slope (red curve below). You will remember that the myoglobin curve rose very sharply on the far left side of the curve which indicated that it has a high affinity for oxygen. Something that shifts the curve to the right and lowers the slope lowers the oxygen affinity. This important phenomenon is known as the Bohr Effect after Christian Bohr made the observation in 1904. It ensures that maximum oxygen is delivered to tissues that are metabolically active, such as exercising muscle. The opposite happens in the lungs when CO2 is lower, the oxygen dissociation curve shifts to the left with increased slope (blue), hemoglobin then having a higher affinity to load up with oxygen.
BPG - 2,3-bisphosphoglycerate
Around 1921, Joseph Barcroft determined that hemoglobin in solution had a higher affinity for oxygen than within red blood cells. Reinhold and Ruth Benesch in 1967 finally determined that the reason for this is due to a substance known as BPG that binds to hemoglobin and decreases its oxygen affinity. Again, this is a phenomenon that increases the ability of hemoglobin to unload oxygen and deliver it to the tissues. This is the reason that myoglobin has a higher oxygen affinity that hemoglobin. It does not bind to BPG. We will discuss why that is in a later unit.
There is another situation where BPG binding to hemoglobin is a benefit - in pregnancy! While fetuses are developing, they are dependent on getting the oxygen they require from the mother's blood. Just like the example of myoglobin having great affinity for oxygen to increase the transfer of oxygen from hemoglobin to myoglobin, a similar situation exists in fetuses. Fetuses produce a special kind of hemoglobin called fetal hemoglobin. This hemoglobin, like myoglobin, has a higher affinity for oxygen than hemoglobin so that oxygen from maternal hemoglobin is able to unload and be loaded onto the fetal hemoglobin. It is also unable to bind BPG strongly. Again, we will explore the structural basis for this soon.
So as you can see, while the physiological basis of how hemoglobin carries oxygen from the lungs to the tissues was fairly well understood around 100 years ago, the structural basis for its function was not elucidated until about 50 years ago. In fact, Max Perutz didn't even solve the initial structure until 1957! Now that we have a basic understanding of the known characteristics of oxygen binding to hemoglobin, let's try to connect it with our understanding of the structure.
In order to understand how oxygen binds to hemoglobin, we have to go back to the allosteric model of hemoglobin that we discussed previously as well as our exploration of oxygen binding to the heme within the globin fold.
You will remember that we discussed how hemoglobin can exist in 2 states - a deoxyhemoglobin state with low oxygen affinity, also known as the T conformation; and an oxyhemoglobin state with higher oxygen affinity, known as the R state. When the first oxygen binds to one of the hemes of hemoglobin, it causes a switch from the T to the R conformation, increasing the "appetite of hemoglobin for oxygen, allowing the 2nd, 3rd, and 4th heme irons to bind oxygen molecules, thereby fully saturating the hemoglobin. The 2 states are the 2 alpha-beta dimers that make up hemoglobin rotating slightly with respect to each other.

How does one oxygen molecule binding to one heme cause a conformational switch of the whole tetramer? That goes back to our demonstration of the structure of the heme group and the protein environment that it is embedded in. Remember, the iron of a heme group is bound by 4 nitrogens in the heme group and one (proximal) histidine from a globin subunit. Without oxygen bound, the heme is in a domed shape with the iron sitting slightly outside the plane of the heme in the direction of the proximal histidine. The 6th coordination position of the iron atom opposite from the proximal histidine is where oxygen binds. When oxygen binds, it pulls the iron into the plane. This starts a chain reaction which is the "switch" from T to R conformations. The oxygen binding pulls the iron into the plane of the heme, which pulls on the proximal histidine, which pulls the F helix that the histidine is a part of as well as the EF and FG segments attached to the F helix. This slight movement disrupts hydrogen bonds between one alpha-beta dimer and the other alpha-beta dimer that stabilizes the T conformation. When that happens, the dimers shift to the R conformation which is stabilized by other hydrogen bonds between the dimers. As this happens, the oxygen affinity for oxygen is increased and the remaining hemes bind O2. The reverse occurs when the first oxygen is pulled off, destabilizing the R form and allowing it to revert back to the T form, releasing the rest of the oxygen molecules.
So finally, how does BPG reduce oxygen affinity of hemoglobin (compared to myoglobin and fetal hemoglobin), as we discussed in the last section? It stabilizes the T conformation making it more difficult to bind oxygen, break the hydrogen bond, and switch to the R conformation. It does this by binding between His2, Lys82, and His143 of each of the 2 beta globins, cross-linking them together. This adds more interactions that need to be broken to make the switch. Once the first oxygen does bind though, it pops the BPG off and the switch is made, allowing oxygen to bind to the other heme groups. Because BPG cross-links across the 2 alpha-beta dimers, a monomer like myoglobin can't bind BPG and so is unaffected by it.
We also learned in the last section that fetal hemoglobin has a higher affinity for oxygen than adult hemoglobin. What is the structural basis for that? It turns out that residue 143 in fetal hemoglobin is an uncharged serine rather than a positively charged histidine that is present in adult hemoglobin. Because of this, BPG does not bind as strongly to fetal hemoglobin as it does to adult and thus has higher oxygen affinity. Isn't that cool?
I know this is a lot to digest. I will show you this on a physical model in a video soon! We will also spend more time exploring fetal hemoglobin when we cover globin genetics in a later unit. But in the meantime, discuss this with your teammates and make sure it makes sense to you. You probably have a lot of questions. In fact, the more you understand this, the more new questions you will probably come up with. That's great! There are all sorts of modeling projects you can explore with these concepts. Building a model is a great way to better understand these complicated structures and relationships. Consider choosing to explore one of these mechanisms for your final modeling project.
Summary of Hemoglobin
3. Globin Genetic Disorders
Up to now, we have been focused on the hemoglobin protein complex (tetramer) of alpha and beta globin subunits. But there are other forms of globins that are expressed at different times during development.
We briefly discussed fetal hemoglobin and how it has a slightly higher oxygen affinity that adult hemoglobin which is important during pregnancy in allowing oxygen to be transferred from the mother's adult hemoglobin to the fetus' hemoglobin. The genetics of hemoglobin are fairly complicated but we need to have a basic understanding of it before we discuss mutations that can lead to hemoglobin disorders, generally referred to as hemoglobinopathies.
Hemoglobin Genes and their RegulationThere are a total of 8 different genes that are involved in the expression of hemoglobin throughout the lifespan of a human! They are depicted by dark blue boxes in the figure below. There are 3 genes on chromosome 16 that are alpha-like. That means that at various times during development, the proteins they encode for have a similar function to the alpha subunit in adult hemoglobin. In fact, there are actually 2 genes that are designated as alpha that are both expressed during adulthood. On chromosome 11, there are 5 more genes that are beta-like and are similarly differentially expressed throughout development. The light blue genes in the figure below are pseudogenes, which are genes that have lost their ability to be expressed over time and so are inactive. The various combinations lead to 3 forms of embryonic hemoglobin, 2 forms of fetal hemoglobin, and 2 forms of adult hemoglobin.
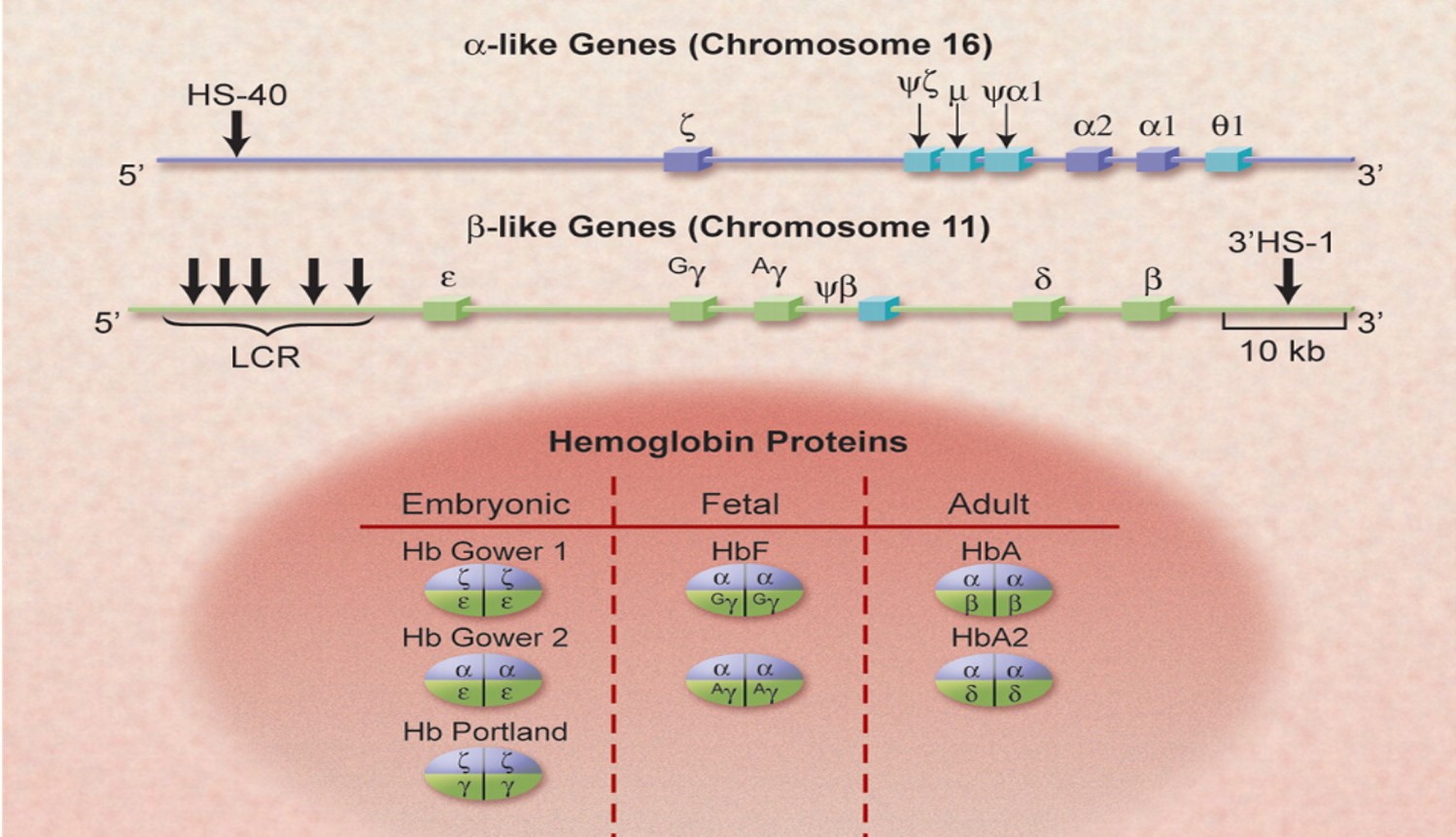
Fetal hemoglobin is made up of 2 alpha globins (the other alpha-like globins are only expressed in the embryo) and, 2 gamma globins, instead of the 2 beta globins found in adults. Near the end of gestation and throughout the first few months, gamma-globin expression decreases and beta globin expression increases. The locus control region (LCR) at the 5' end of the cluster of beta-like genes is responsible for the developmental switching of expression (see the graph below). Isn't that interesting? Remember this nugget later when we get to hemoglobin disorders and therapies...
This very complicated but amazing system has come into existence over millions of years of gene duplication and evolution. There is much to explore here but it is beyond the scope of this part of the program to go much further. If you are really interested in this, consider exploring hemoglobin evolution or transcriptional regulation for your molecular story.
The figures above came from the attached review that I really like. It is very comprehensive so read it or parts of it if you wish. It is chock full of potential project ideas!
Hemoglobin research and the origins of molecular medicinePart One
Now that we understand how the hemoglobin gene family is arranged on chromosome 16 and 11, let's zoom in on a portion of the beta-globin gene on chromosome 11 with the Beta-globin Gene Map, if you have one available.
Take some time examining the student version of the map. What do you think the red and black lines of letters represent? What do you think the 3 lines of blue letters represent? Why are there 3 of them? What do the asterisks mean? Why does the numbering start at 61981? What might that signify (think about what you learned in the last section)? Do you have any other questions about what these sequences represent? Discuss these questions with your team and then return for more instructions!
Next, print out the beta globin sequence below and assemble it. Look at the sequence you annotated in the Globin Structure section and highlight the alpha helices and circle the proximal and distal histidines on the newly assembled sequence. Use the protein sequence to find the entire sequence on the gene map. This might take some time. When you are done, return to Part Two for more instructions.
Expanded Human Betaglobin Sequence
Part Two
Before we discuss your findings, we need to define what the red, black, and blue lines signify. The black line represents what is called the template strand and the red line represents the complementary non-template strand. It is the strand that is "read" by RNA polymerase to transcribe the template to mRNA. That mRNA looks a lot like the red strand, complementary strand except the T's would be U's in RNA. This is the sequence that will be translated into the beta globin protein. There is another difference between the mRNA sequence and the red DNA sequence shown but we will discuss that later. The blue lines of letters represent the theoretical amino acid sequences if the codons were read in each of 3 different reading frames. That is, if you start with the first 3 nucleotides of the red sequence and read the triplet codons in order, you will generate the top blue sequence. If you skip the first nucleotide and begin to read triplet codons from the 2nd nucleotide on, you will generate the 2nd sequence and the 3rd is generated by beginning with the 3rd nucleotide. Thus all 3 possible reading frames are represented. If you begin at #4, it will be in frame with the first one.
If you were successful in finding the entire protein sequence within the blue lines, you will have realize that the sequence was broken into 3 pieces. The 3 pieces happened to be on 3 different blue amino acid sequences and it should have jumped between the sequence as shown below. Why might it do that? Why are there nucleotides between the 3 pieces? Does the ribosome have to keep jumping around on the mRNA to make a protein?
Earlier it was mentioned that there was another way that the mRNA would be different from the non-template (red) DNA. The answer is called splicing. Our DNA has regions of nucleotide sequence within a gene that codes for protein (exons) and interspersed nucleotides that don't (introns). When the RNA is first being transcribed, it must undergo processing before it becomes a messenger RNA that can be read by a ribosome to make a protein. One of those types of processing is called splicing. That is when the introns are taken out and the exons are connected so that the correct coding sequence reads in one continuous reading frame. Why do introns exist? Can you speculate why this system might be an advantage to an organism?

Where did the exons begin and end compared to the protein sequence and known structure? Which exon encodes for the globin fold? You should have determined that the asterisks signify stop codons. Why do you think the sequence starts at 61,981? The sequence that comes before the beta-globin gene contains the other beta-like genes we mentioned earlier (see section on Hemoglobin Genes and their Regulation) and the Locus Control Region (LCR) that is responsible for switching expression of the various genes on and off. Again, very cool topic that is too deep to cover here but keep it in mind for a project!
For more information, take a look now at the teacher version of the beta-globin gene map. You might want to use the teacher version again as you work through the Disorders section as it contains a wealth of information regarding mutations.
This is a great resource for understanding the science behind sickle cell disease as well as the personal struggles that patients go through on a daily basis.
Process of Discovery of Sickle Cell Anemia
This activity will lead you through the experiments done in the 1950's that led to the understanding of the molecular basis for Sickle Cell Disease.
Possible Sickle Cell "Cures"
The Scientific American article below discusses a girl with 2-S globin alleles (beta globin with the sickle mutation) but does not have any symptoms because of another mutation that allows her body to keep producing reasonable quantities of the fetal isoform of beta globin, gamma globin. This is called "Inherited Persistance of Fetal Hemoglobin".
4. Defining Your Globin Protein Story
Now that you are armed with an extensive understanding of what globins are, how the globin fold is important to their unique structure, and how their structure enables their critical function, it is time to decide what specific globin story you would like to explore.
The expandable content below will give you some suggested topics you may choose to focus on, recommended globin research papers and reviews to read, and link to a collection of globin structure files to use when designing your 3D printed protein model.
Globin Topics and Review ArticlesDo you want to find out more about fetal hemoglobin and how it compares to adult hemoglobin? Are you interested in the genetic cause of Sickle Cell Anemia and some of the potential cures? Do you think earth worm hemoglobin might be fun to model? . . . There is an endless list to globin-related questions that you can ask and then explore.
Look over the Topic Lists below (not comprehensive) as well as the Review Papers to get an idea of which direction you want to go for your unique globin protein story.
Research Topics
- Lamprey oldest form of hemoglobin, dimer, not tetramer, weakly cooperative. What is known about its structure-function?
- Myoglobin, is it only for diving mammals or is it important for humans? It's nice, but can we live without it?
- Compounds that affect the affinity of hemoglobin for oxygen might be used for good (treat disease) or evil (blood-doping in athletics). How do some of them work?
- BPG levels in the red blood cells can be regulated due to different conditions or stresses. It has also been implicated in some diseases.
- There are lesser understood globins called cytoglobin and neuroglobin. What do they do? How do they differ from myoglobin?
- Genome editing has recently been used as a potential treatment for sickle cell and beta-thalassemia. What is the strategy?
- Carbon monoxide binds to hemoglobin extremely tightly, blocking the binding of oxygen, which is why it is so deadly. How does that work?
- Nitric oxide is a gas that is an important modulator of blood vessel function and has been shown to bind to and be transported by hemoglobin. How does that work?
- Heterozygosity for sickle cell anemia or thalassemia (meaning a person has on normal copy of the beta globin gene and one mutated copy) confers protection from malaria. How does that work?
- Using the CRISPR system to cure Sickle Cell disease.
Globin Papers
Below are PDFs of some nice reviews of diseases and disorders related to globin function/dysfunction. Explore these and other sources to decide what you might want to choose for your protein story. Some journals have devoted special issues to globins and include articles (with links to pdfs) on lots of different aspects of globin biology.
Papers on Neuroglobin
- Brain globins in physiology and pathology
- The Anti-Apoptotic Role of Neuroglobin
- Neuroglobin: From structure to function in health and disease
- Critical re-evaluation of neuroglobin expression reveals conserved patterns among mammals
- Structure and Reactivity of Hexacoordinate Hemoglobins
- Human Brain Neuroglobin Structure Reveals a Distinct Mode of Controlling Oxygen Affinity
- The crystal structure of wild-type human brain neuroglobin reveals flexibility of the disulfide bond that regulates oxygen affinity
- The binding of cytochrome c to neuroglobin: A docking and surface plasmon resonance study
- Neuroglobin, a Factor Playing for Nerve Cell Survival
Papers on Globin Evolution
- Globin Evolution Review
- How Different Amino Acid Sequences Determine Similar Protein Structures : The Structure and Evolutionary Dynamics of the Globins
Papers on CRISPR Cas9 and Sickle Cell
Defining Your Globin Story
Your MAPS Team will want to have a clear idea of what your globin story is before you start your model design. And one of the best ways to define your protein story is to write a scientific abstract.
Abstracts are, by definition, short. You might want to start with one sentence for each of the sections listed below, and then add additional sentences as needed.
- Introduction: What is the overall relevance of your protein of interest?
- Question: What is the particular story of your protein that your team will model?
- Findings: What answers to your question were you able to determine through your modeling project? Be specific as to what structural elements your team has modeled and how they are relevant to your story.
- Conclusion: How do your findings relate to the big picture? What important questions related to your protein story are still being actively researched?
Once you have defined the globin protein story you want to tell, you will need to find a protein structure file that you can work with in Jmol to design your 3D printed globin model.
Designing and building your own physical globin model will give you great insight into specific structures that are important to the globin protein story you have chosen to focus on. Even if you don't have your design 3D printed, studying the 3D design in specialized computer software is still helpful.
Below are a few additional links that may help you in your globin structure search.
What Does it Mean to "Design" a Protein Model?
Tim Herman, PhD Explains How to Approach Your Model Design in Jmol
"Designing" a protein model means you explore a protein structure in Jmol, and then simplify the way the protein is visually displayed to make the key features of the protein that help communicate your molecular story more obvious.
This can mean hiding some atoms that are not important for your protein story, changing the display format of certain parts of your protein structure or changing colors to best highlight the most important parts of the protein structures. In the next section, you will learn the Jmol commands needed to accomplish this.
How to Approach Your Model Design
Tim Herman, PhD Explains How to Approach Your Model Design in Jmol

All 3D printed protein models made in the MAPS program are designed using the program Jmol. You start with a protein structure file (.PDB file), which contains the 3D locations of all of the atoms that make up the protein. Using the Jmol commands you will learn below, you can then edit the way the protein is displayed, hiding some atoms, changing display formats and customizing colors. When happy with your design, you can export your protein model from Jmol in file formats suitable for 3D printing.
We have created a detailed Jmol Training Guide that will cover everything you need to know to design and build your model. The Jmol Training Guide is broken down into four main sections, which we strongly recommend you explore in order until you are comfortable with Jmol and designing protein models for 3D printing.
- 1) Getting Started:
- Accessing Jmol
- Loading Structures
- Saving and Reloading - 2) Simplifying Your Protein:
- Exploring the Structure
- Focusing on the Backbone - 3) Adding the Details:
- Adding Sidechains
- Adding Ligands
- Adding Molecular Bonds - 4) 3D Printing Your Design:
- Adding Support Struts
- Checking Your Design
- Print it Yourself
- Pay Someone Else to Print it
Moving to the Next Phase
Once your team has finished the protein story phase, move on the Capstone Experience, where you will learn how to present your project using your 3D printed model and a variety of complimentary digital and print media.