The 2016 Science Olympiad Protein Modeling Event
Proteins Involved in the Biosynthesis and Subsequent Signaling of the Neurotransmitters Dopamine and Serotonin
Inspiration for the theme: Sepiapterin Reductase and the Beery Twins
 Be extra sure to explore the information in all three tabs below
Be extra sure to explore the information in all three tabs below
Introduction
In the video below, Tim Herman, Ph.D. from the MSOE Center for BioMolecular Modeling provides an introduction to this year's topic, Sepiapterin Reductase and the Beery Twins.
The Beery family story is a genomics story. It is an example of how whole genome sequencing was used to arrive at a definitive, molecular diagnosis of the twins’ medical condition.
Your primary resource in exploring this story will be the original research paper published by the research group at the Baylor College of Medicine who sequenced the twins’ genome and then identified mutations in the sepiapterin reductase gene as the cause of their dystonia disorder. A copy of that paper is provided here:
Paper Abstract:
Whole-genome sequencing of patient DNA can facilitate diagnosis of a disease, but its potential for guiding treatment has been under-realized. We interrogated the complete genome sequences of a 14-year-old fraternal twin pair diagnosed with dopa (3,4-dihydroxyphenylalanine)–responsive dystonia (DRD; Mendelian Inheritance in Man #128230). DRD is a genetically heterogeneous and clinically complex movement disorder that is usually treated with L-dopa, a precursor of the neurotransmitter dopamine. Whole-genome sequencing identified compound heterozygous mutations in the SPR gene encoding sepiapterin reductase. Disruption of SPR causes a decrease in tetrahydrobiopterin, a cofactor required for the hydroxylase enzymes that synthesize the neurotransmitters dopamine and serotonin. Supplementation of L-dopa therapy with 5-hydroxytryptophan, a serotonin precursor, resulted in clinical improvements in both twins.
Many of the figures from the Lupski and Gibbs paper are discussed below, along with other resources and background material you will need as you explore the Beery family story.
Table of Contents:
- The Beery Family
- The Beery Family History
- The Beery Family Pedigree
- The Genetic Basis of the Beery Twins' DOPA-Responsive Dystonia
- Genome Sequencing of the Beery Twins
- Validation of Two Sepiapterin Reductase Alleles by Sanger DNA Sequencing
- The Full Sepiapterin Reductase Gene
- The Role of Sepiapterin Reductase in
the Biosynthesis of Two Neurotransmitters
- Neurons and the Synaptic Cleft
- Sepiapterin Reductase - What Does It Do?
- What does the cofactor BH4 have to do with the Beery family story?
- A Summary of Serotonin and Dopamine Biosynthesis
- Mapping the Mutations to a
Physical Model of Sepiapterin Reductase
- The Robert Huber Paper
- The Locations of Retta and Joe Beery's Mutations
- The Solution
- But Life is Complicated
- The Beery Family Today
The Beery Family
The Beery Family History
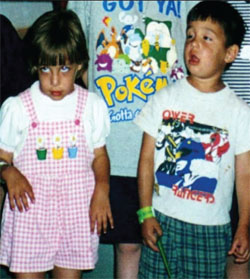
The Beery Twins, Noah
and Alexis, as Children
Soon after Alexis and Noah Beery were born it was obvious to their parents, Joe and Retta, that the twins were different from their older brother, Zach. They demonstrated poor muscle tone, cried nonstop, vomited frequently and missed developmental milestones. Physicians diagnosed the twins with cerebral palsy. By age 5, Alexis was having difficulty swallowing and was wasting away, symptoms not consistent with cerebral palsy. Retta came across an article about a rare disorder, dopa-responsive dystonia (DRD), which is caused by a deficiency of the brain neurotransmitter dopamine. The symptoms matched those of her daughter. Shortly after, both twins were treated with L-DOPA (dopamine precursor), and their condition improved dramatically. However, as the twins grew older, it became apparent that even this diagnosis was incorrect as their health began to further deteriorate.
To learn more about the Beery family and their journy, explore their website at http://dystonia.thebeerys.com.
The Beery Family Pedigree
Examination of the Beery family pedigree shows a history of depression on the paternal side and a history of fibromyalgia and undisclosed neurological disorder on the maternal side, in addition to DRD in the twins.
The Genetic Basis of the Beery Twins'
DOPA-Responsive Dystonia
Genome Sequencing of the Beery Twins
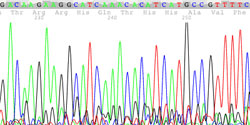
An Example of Genome Sequencing Data
In the case of the Beery twins, whole genome sequencing led to a more complete understanding of the molecular basis of their disease and informed a change in their medical treatment. Joe Beery, Alexis and Noah's father, was working as the VP for Information Technology at Life Technologies (a biotech company involved in NextGen DNA sequencing) and arranged to have the twins' genomes sequenced by a group at the Baylor College of Medicine. More than 2 million variations were found in the DNA sequence of the twins' genomes, compared to a reference human genome. This level of variation is quite normal. Various bioinformatics filters were applied to eliminate variants that were not thought to be responsible for their condition. Following this filtering, only three variants remained as candidate genes. One of these was sepiapterin reductase (SPR). The nucleotide sequence of the twins' SPR genes revealed two different mutations – each of which were believed to result in an inactive enzyme.
Validation of Two Sepiapterin Reductase Alleles by Sanger DNA Sequencing
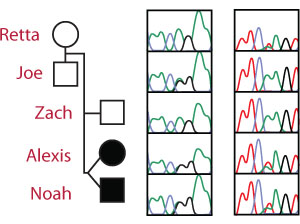
The SPR Genotype Sequencing Traces
Recent advances in DNA sequencing technology have made it possible to rapidly sequence whole human genomes at a cost approaching $1,000. Although NextGen DNA sequencing is rapid and inexpensive, it is not as accurate as other sequencing methods. Therefore, Sanger DNA sequencing was used to confirm the sequence of the twin’s SPR genes.
The Sanger sequencing traces (right) show the SPR genotype for each member of the Beery family.
The Arg150Gly mutation is an A → G mutation on chromosome 2 at nucleotide 72,969,094 leading to the replacement of Arginine with Glycine. The unaffected father is heterozygous (A/G) for the pathogenic Arg150Gly allele at the first locus and homozygous (A/A) for the wild-type allele at the second locus.
The Lys251X mutation is an A → T mutation on chromosome 2 at nucleotide 72,972,139 resulting in the conversion of a Lysine codon (AAG) to a STOP codon (UAG). The unaffected mother is homozygous (A/A) for the wild-type allele at the first locus but heterozygous (A/T) for the pathogenic Lys251X allele at the second locus.
Each affected twin is a compound heterozygote - (A/G and A/T) with a different pathogenic mutation on each allele.
The Full Sepiapterin Reductase Gene
Explore the sepiapterin reductase gene map shown below to see Retta Beery's mutation (highlihgted in pink) and Joe Beery's mutation (highlihgted in blue). Click Here to download the entire sepiapterin reductase gene as a PDF document.

The Role of Sepiapterin Reductase in
the Biosynthesis of Two Neurotransmitters
Neurons and the Synaptic Cleft
Sepiapterin reductase is an enzyme associated with the biosynthesis of two neurotransmitters, dopamine and serotonin. But to understand the specific role of sepiapterin reductase in our nervous system, an overview of neuron structure and the synaptic cleft is needed. In the two videos below, Gina Vogt from the MSOE Center for BioMolecular Modeling discusses the shape of neurons, the location of the synaptic cleft and the role of dopamine and other neurotransmitters in the neurotransmission.
Sepiapterin Reductase - What Does it Do?
Sepiapterin reductase is an enzyme that is responsible for the biosynthesis of a cofactor known as tetrahydrobiopterin (BH4).
Enzyme cofactors are small molecules that enable some enzymes to catalyze difficult reactions that would otherwise be impossible with only the twenty amino acid sidechains. In other words. . . most enzymes contain "active sites" that bind specific substrates in such a way that several active site amino acid sidechains are precisely positioned to perform a chemical reaction. Sometimes these active site sidechains require the help of cofactors to create the necessary chemistry to catalyze the reaction.
What does the cofactor BH4 have to do with the Beery family story?
The cofactor BH4 is needed by an enzyme that participates in the biosynthetic pathway that converts the amino acid tyrosine into the neurotransmitter dopamine. A similar enzyme uses BH4 to convert tryptophan into serotonin. Both of these enzymes are "hydroxylases" that add a hydroxyl group to an amino acid.
In the video below, Tim Herman, Ph.D. from the MSOE Center for BioMolecular Modeling provides an overview of neurotransmitter biosynthesis and the role sepiapterin reductase plays in the pathway.
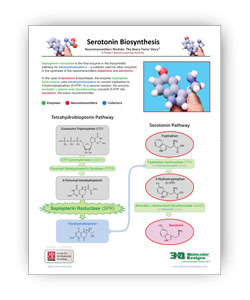
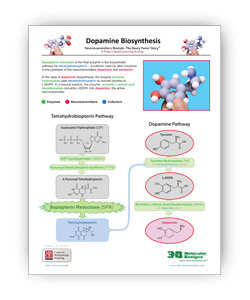
A Summary of Serotonin and Dopamine Biosynthesis
The two .PDF links below summarize the steps needed to turn tryptophan into serotonin and tyrosine into dopamine.
Sepiapterin reductase is the final enzyme in the biosynthetic pathway for tetrahydrobiopterin – a cofactor used by other enzymes in the synthesis of the neurotransmitters dopamine and serotonin.
In the case of dopamine biosynthesis, the enzyme tyrosine hydroxylase uses tetrahydrobiopterin as a cofactor to convert tyrosine to L-DOPA. In the case of serotonin, the enzyme tryptophan hydroxylase uses tetrahydrobiopterin to convert tryptophan to 5-hydroxy tryptophan (5-HTP). In a second reaction in each pathway, aromatic L-amino acid decarboxylase (AAAD) converts both L-DOPA to dopamine and 5-HTP to serotonin, the active neurotransmitters.
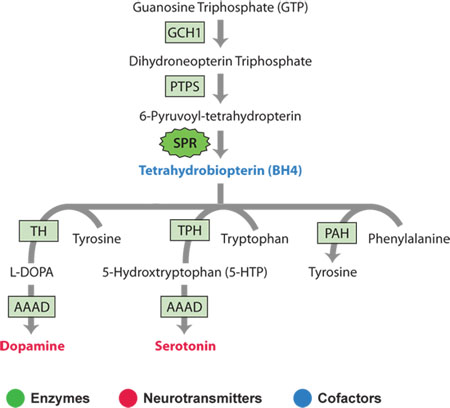
Click here for expanded definitions of Enzymes, Neurotransmitters and Cofactors
The two .PDF links below summarize the two pathways involved in producing tetrahydrobiopterin and neurotransmitters.
Mapping the Mutations to a
Physical Model of Sepiapterin Reductase
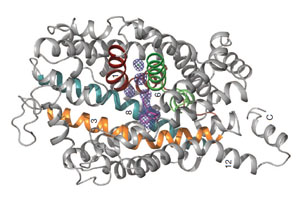
The Robert Huber Paper
The 3-dimensional structure of a mouse sepiapterin reductase was first determined by Robert Huber’s group in 1997. An annotated version of the "primary citation" (the original research paper that reported this structure) is provided below. This is a very detailed research paper. You are not expected to read and understand the entire paper. To guide your reading, we have highlighted those sections of the paper that will provide you with a general introduction the structure and function of the protein.
Note that your pre-build model is based on the structure of a mouse sepiapterin reductase protein, as described by 1sep.pdb. We chose the mouse enzyme for this protein modeling event because:
- It was the first structure of this protein that was solved (in 1999).
- Tetrahydrobiopterin was bound in the active site of this enzyme.
- The mouse enzyme is 74% identical (and 88% similar) to the human enzyme.
The Locations of Retta and Joe Beery's Mutations
As you prepare for this protein modeling event, you should determine the molecular consequence of the two mutations identified in the twins’ SRP gene – and map these changes onto the 3D structure of the sepiapterin protein.
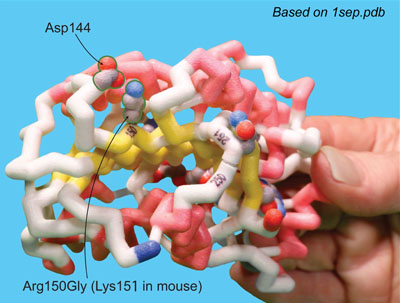
Joe Beery's Mutation, an Arginine to a Glycine
Joe Beery’s Arg150Gly missense mutation (left image) results in a change from a positively charged basic arginine amino acid at position 150 to an uncharged glycine. This would disrupt a salt bridge interaction with the negatively charged aspartic acid at 144 in the functional enzyme.

Retta Beery's Mutation, a Lysine to a STOP Codon
Retta Beery’s Lys251X nonsense mutation (right image) results in a change from lysine at position 251 to a premature STOP codon, causing truncation of the enzyme.
The Solution
If Alexis and Noah can’t make enough BH4 to support normal levels of dopamine and serotonin, how can we fix this problem?
The solution is to provide the neurotransmitter precursor immediately downstream from the biosynthetic step that requires BH4 – namely L-DOPA and 5-hydroxy tryptophan. Daily doses of these two precursors allow Alexis and Noah to lead the lives of normal, healthy teenagers.
But Life is Complicated
As one final twist in this story, the L-DOPA and 5-hyroxy tryptophan medication that Alexis and Noah take is also supplemented with another small molecule called carbidopa. Can you figure out what carbidopa is and what role it plays in the twins' medication?
The Beery Family Today

The Beery Twins Today
Noah and Alexis have a passion to share their story. According to a recent blog post, "It's our hope and prayer that people will use their voices in their medical care or any other area of their lives, and never, ever give up hope - no matter how dire the situation may seem. Because hope is unlimited and has no boundaries!"
In order to truly appreciate and successfully model this year's Protein Modeling Event structures, a thorough understanding of proteins, DNA and genomes is needed. This section will explore these amazing macromolecules in more detail using suggested physical model kits, online resources and additional websites.
Protein Structure
Proteins are long linear sequences of amino acids
that fold into complex 3-dimensional shapes
following basic principles of chemsitry.
In this first of two videos on protein folding, Tim Herman, Ph.D. from the MSOE Center for BioMolecular Modeling uses the Water Cup from 3D Molecular Designs to demonstrate some basic principles of chemistry.
In this second of two videos on protein folding, Tim Herman, Ph.D. from the MSOE Center for BioMolecular Modeling uses the Amino Acid Starter Kit from 3D Molecular Designs to demonstrate how the basic principles of chemistry directly affect protein folding.
In this video, Tim Herman, Ph.D. from the MSOE Center for BioMolecular Modeling uses the Alpha-helix Beta-sheet Construction Kit from 3D Molecular Designs to demonstrate the form and function of secondary structures in proteins.

Protein Databank (www.rcsb.org) Resources
Learn about protein structure and function with this overview printout and video developed by the RCSB Protein Databank.
- What is a Protein? (PDF)
- What is a Protein? (video)

Protein Structure Jmol Tutorials
The Protein Structure Jmol Tutorials walk through the four levels of protein structure using interactive Jmol molecular visualizations, including real protein examples with interactive controls.
- Primary Structure (Webpage)
- Secondary Structure (Webpage)
- Tertiary Structure (Webpage)
- Quaternary Structure (Webpage)

3D Molecular Designs Educational Kits
These engaging, hands-on kits make learning protein structure basics easy. Users will fold a protein while exploring how the chemical properties of amino acids determine its final structure.
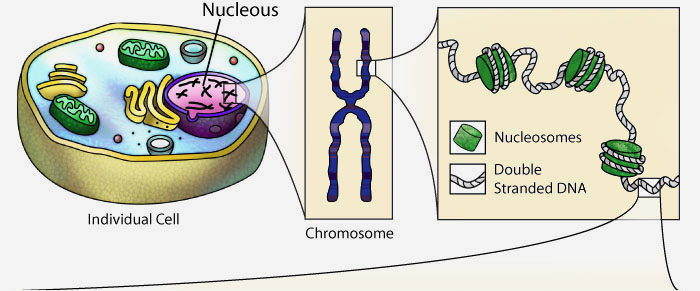
DNA and the Flow of Genetic Information
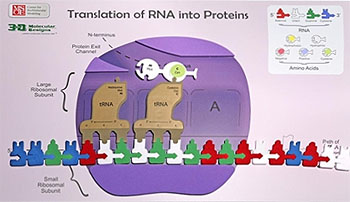
Every cell in your body remembers how to accurately make thousands of different types of proteins. They store the instructions for each protein's primary structure (sequence of amino acids) in the form of DNA.
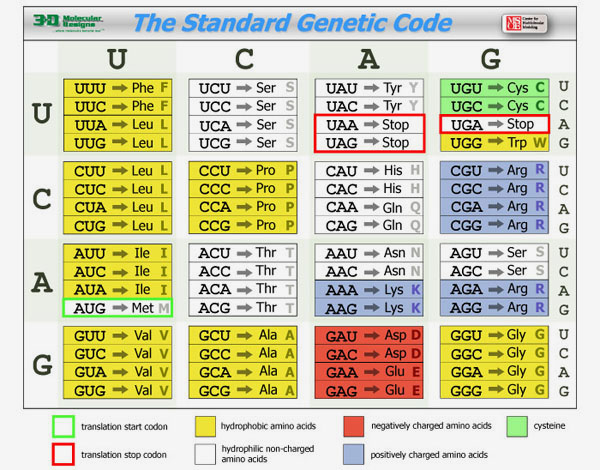
When a cell needs a new copy of a protein, it accesses this DNA, creating a copy of the specific instructions (called a gene) for the desired protein. This copy, made of RNA, is then interpreted by a molecular machine called a ribosome. Ribosomes use the instructions stored in the RNA to link amino acids together in the correct sequence.
The newly synthesized protein sequence then folds following basic principles of chemistry into a complex 3-dimensional shape, giving it a specific form and function.
Click here for a more detailed review of the flow of genetic informationGenomes and Personalize Medicine

Personal Genome Analysis
This excerpt from the 2013 HudsonAlpha Biotechnology Guide Book provides an overview of personal genome analysis, covering recent technological advances in the feild and how genome data is used.
Personalized Medicine
This excerpt from the 2013 HudsonAlpha Biotechnology Guide Book provides an overview of personalized medicine, describing how genome sequencing is being used by doctors to provide unique and individualized medical care to patients.

Diagnostic Clinical Genome and Exome Sequencing
This article from the New England Journal of Medicine discusses using genome and exome sequencing to diagnose clinical disorders.
This Year's Proteins

The table below lists the proteins that will be featured as the 2016 pre-build as well as the various on-site builds at invitational, regional, state and national competitions.
Be Sure to Click on Each Protein Listed Below to reveal additional resources that you should explore as you prepare for each level of competition. In addition to providing the pdb file for the protein that will be featured, these links also provide a copy of the original research paper (the "primary citation") that reported the protein's structure. Questions regarding the paper and the structure presented in the PDB file will be asked on the exam you will complete at each competition.
Pre-Build Model Sepiapterin Reductase 1sep.pdb Invitational On-site Model G-Protein 1gia.pdb Regional On-site Model DOPA Decarboxylase 1js3.pdb State On-site Model Dopamine Transporter 4m48.pdb National On-site Model Dopamine Receptor 3pbl.pdb