



This tutorial was created with funding from NSF-DUE (1022793, 1323414, 1725940) for the CREST program. Last revision 3/2021
Alkaline phosphatases are found in all living things and are characterized by their ability to hydrolyze phosphate groups from a wide range of substrates in alkaline conditions. Alkaline phosphatases function as homodimers, consisting of two identical subunits called monomers. Each monomer has an active site with three metal ions, typically two Zn2+ and one Mg2+ ions. These metals play key roles in binding substrates with terminal phosphate groups. Although alkaline phosphatases share these common features, there are differences among the various enzymes in terms of their activity and thermal stability.
The structure of alkaline phosphatase from bacterial species, especially that of E. coli, has been extensively studied, and numerous structures are available from the Protein Data Bank (PDB). These structures were filed into the PDB by research scientists who crystallized alkaline phosphatase enzymes in various states (i.e. incorporating a variety of ligands and/or mutations in active site residues). All of the protein structure files uploaded to the PDB provide insight into the catalysis of phosphate-containing substrates by alkaline phosphatase.
Although bacterial alkaline phosphatases (EAP; "E. coli alkaline phsophatase") have some primary amino acid sequence homology with mammalian enzymes, there are some striking differences. For example, some free L-amino acids serve as uncompetitive inhibitors of mammalian, but not bacterial, enzymes. A few structures of human placental alkaline phosphatase (hPLAP) have been determined, and the first intestinal alkaline phosphatase structure from rat (rIAP) became available in the PDB in October 2013.
Undergraduate labs often utilize bovine intestinal alkaline phosphatase (bIAP) to study enzyme kinetics. There are currently no crystal structures of the bovine phosphatase in the PDB. We can, however, make reasonable hypotheses about bIAP structure and function by studying how similar and different it is from other alkaline phosphatases which have been crystallized. This tutorial will allow you to explore and compare the structures of rIAP, hPLAP and EAP. These comparisons will allow you to determine which alkaline phosphatase residues, secondary and tertiary structures are similar (i.e., highly conserved) and where they are different.
Throughout the tutorial, the Jmol image on the top right of the screen will display rIAP. The lower right Jmol image will display either hPLAP or EAP, depending on which button is clicked to launch the tutorial.
Some standard conventions are used throughout the tutorial. Protein structures are displayed in an alpha carbon backbone format. A line (backbone) joins the alpha carbon (to which the sidechain, also known as a residue, is attached) of adjacent amino acids. Sidechains or sugar molecules, when displayed, are colored in standard cpk colors (grey is carbon, red is oxygen, blue is nitrogen, yellow is sulfur, green is zinc, and, in our rendering, firebrick red is magnesium). Any ligands interacting with the protein are colored in "light cpk" - so pink instead of red, etc.
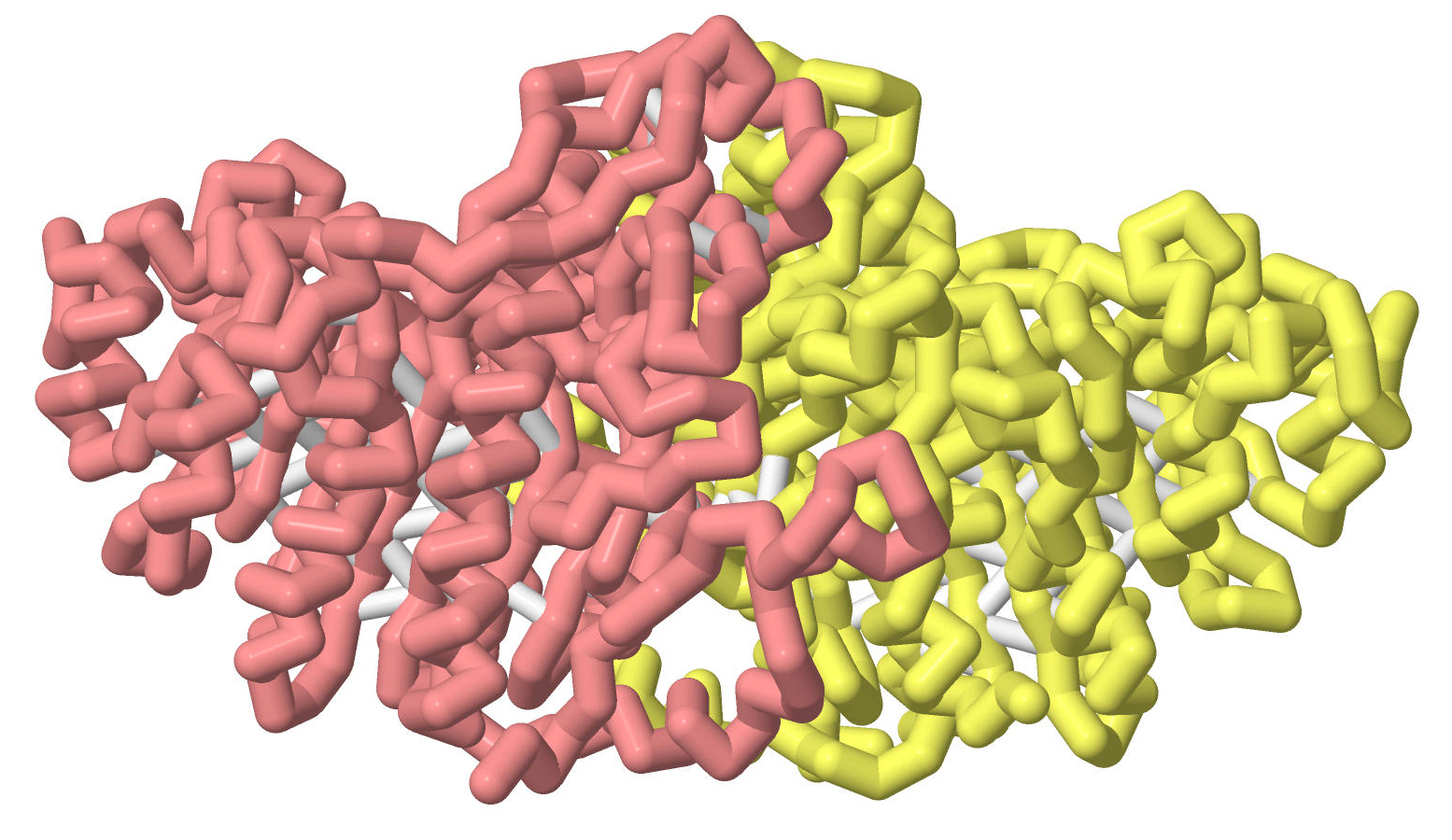
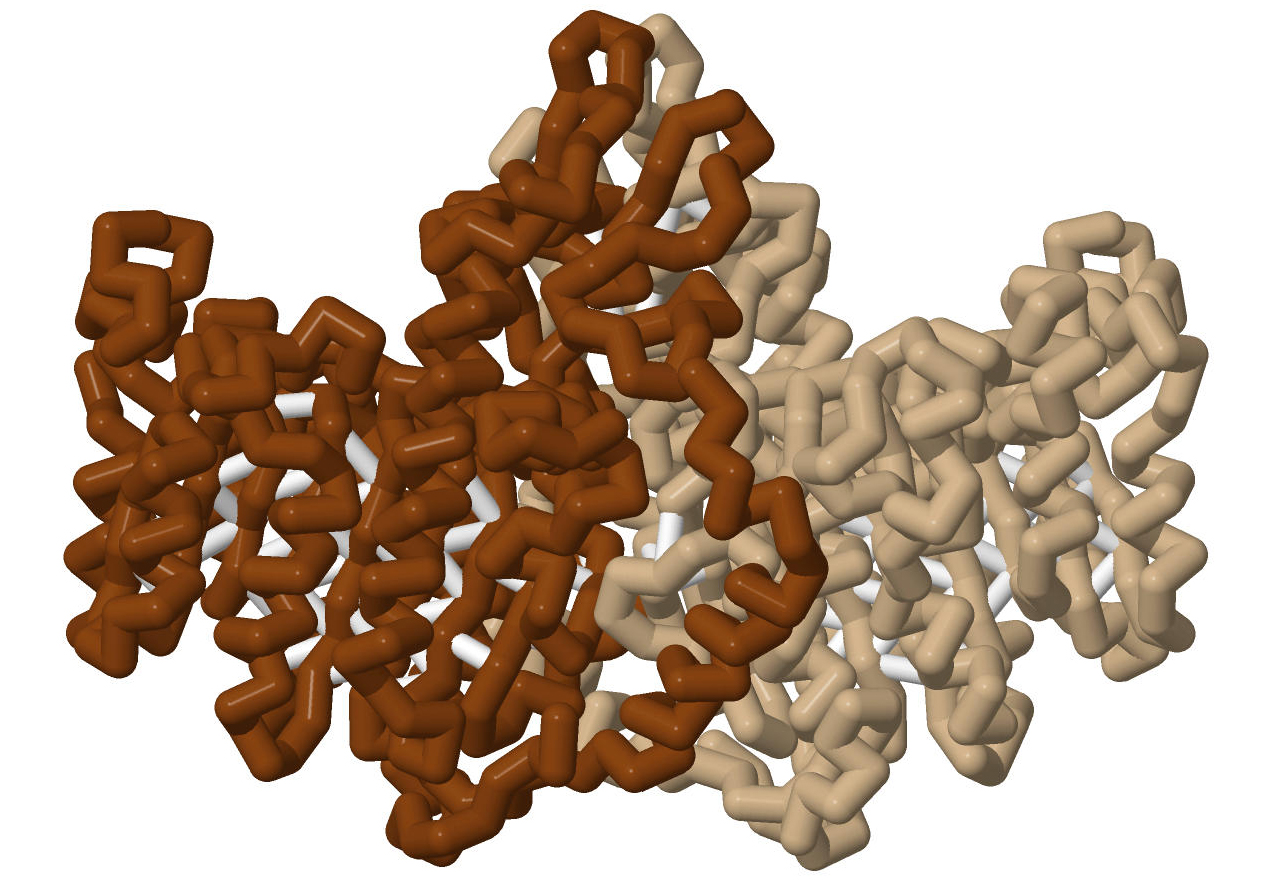
Although all alkaline phosphatases are homodimers, the PDB files for hPLAP display only the monomers. EAP and rIAP are displayed as dimers, but, when compared with hPLAP, only a monomer is displayed.
Structures are aligned when the files are loaded, but can be rotated (click and drag the mouse on the image) or zoomed (shift click and drag the mouse) individually. If you wish to realign, simply click the button to reload the images.
In the Milwaukee School of Engineering's Center for BioMolecular Modeling (CBM), we affectionately refer to the alkaline phosphatase structure as "the bat", though you might also see it as a butterfly with antennae. One of the features of mammalian phosphatases is the presence of a crown domain (the "head" of the bat). Interestingly, when rIAP is expressed in E. coli, the crown domain is disordered and can't be resolved in the structure (not shown, but see 4KJD.pdb). The crown domain is highlighted in light green in these renderings. Depending on the size of your browser window, you may need to zoom in or out (shift left click and drag, or center mouse roller) to see the entire structure. Use the left mouse click and drag to rotate the structures. To return to the same orientation, just click the button to reload.
rIAP and EAP; crown domainMammalian alkaline phosphatases are glycosylated. The rIAP structure has three glycosylation sites.
rIAP and EAP; glycosylation sites on rIAPIt may be challenging to explore the overall structural homology between the two structures, but we will begin by exploring secondary structures separately. In this rendering, we have displayed the beta sheets in rainbow hues. Note that because the scheme applies to BOTH helices and sheets, not all hues will be seen when displaying only helices or sheets.

The alpha helices are mostly oriented in a single, parallel direction, much like a series of pillars. These alpha helices have also been colored in rainbow hues from purples through pinks. Note however the 2 dark blue alpha helices near the interface of the two dimers: how are they oriented relative to most of the other alpha helices in each monomer?
rIAP and EAP helices (dimer)The next view shows both the alpha helices and beta sheets. Which of these secondary structures is found mainly in the interior of the homodimer?
rIAP and EAP helices and sheets (dimer)This image displays the alpha helices and beta sheets in rainbow hues, and the loops connecting these secondary structure are colored a single gray color.
rIAP and EAP backbone - rainbowCrystal structures of human placental alkaline phosphatase (hPLAP) only show the monomeric form. In comparing this structure with rIAP, only the monomer of rIAP is displayed in the upper right window. In this first view, the crown domain is highlighted.
rIAP and hPLAP - crown domainhPLAP has two glycosylation sites, at Asn122 and Asn249. rIAP has three glycosylation sites, as Asn122, Asn281 and Asn408.
rIAP and hPLAP glycosylation sitesThe following four views compare the secondary structure of intestinal and placental phosphatases using the rainbow color scheme. Purple and blue are at the N terminus, red and pink at the C terminus. Loops are colored a single color for each chain.
rIAP and hPLAP sheets (monomer)Both EAP and rIAP bind two Zn2+ and one Mg2+ions in each of the two active sites. The following views explore the amino acids and water molecules that interact with these metal ions in the active site. Each image will begin with a view of the entire alkaline phosphatase, and then zoom into the active site. (Ultimately all structures except for the metal(s), water molecules, and interacting residues will disappear.) Remember to shift click and drag your cursor to zoom in and out.
Zinc metals are shown as dark green balls, and the magnesium metal is shown as a firebrick red. Pause your cursor over each structure: a pop up window will appear and identify the metal, residue, or water molecule you have selected. How similar are the metal-interacting residues in the rat intestinal alkaline phosphatase and E. coli alkaline phosphatase?
Zn1 from rIAP and EAPHuman placental alkaline phosphatase also has two Zn2+ and one Mg2+ions. The following views compare the binding sites of the two mammalian phosphatases.
Zn1 from rIAP and hPLAPThe AP structures in the PDB are from a variety of organisms (mostly prokaryotic), and crystals were created in the presence of a range of substrates and inhibitors. To complicate matters, expressing the same sequence in different organisms yields different structures (compare, for instance, 4KJG with 4KJD), and mutations have been introduced to a number of AP sequences to elucidate important residues and/or to capture the enzyme-substrate complex and reveal the reaction mechanism. For any PDB structure that you view, always keep in mind that the images you see were captured under carefully controlled and unique in vitro experimental conditions. We must always remain aware of the assumptions we make about in vivo macromolecular structure and function, and how evolutionary selection pressures have influenced both.
The following images explore three hPLAP structures determined by a single lab. This will serve as in introduction to the features you may wish to identify as you explore other AP structures.
In each view, a monomer of rIAP is depicted in the top screen, and one of the three hPLAP structures is in the bottom screen. rIAP, based on 4KJG.pdb, is depicted with a brown backbone in the top window for comparison with the three hPLAP structures. Each of these images incorporates a standard coloring scheme, with the backbone displayed as a solid color. Important residues are depicted in cpk coloring, and ligands are in light cpk. Cysteines that form disulfide bonds are displayed in ball and stick and colored in cpk. The backbone of displayed amino acids is colored to reflect its function:
Para-nitrophenol is bound in the active site pocket.
The first hPLAP image (3MK0.pdb) has a light purple backbone and shows para-nitrophenol bound, not at the active site, but at a peripheral site. The active site of this structure has no bound ligands. Zoom in on the empty hPLAP active site and note the orientation of key residues and metals. How do they compare to the rIAP active site?
4KJG and 3MK0The second hPLAP image (3MK1.pdb) has a medium purple backbone. Three molecules of para-nitrophenol are present. These are found at the active site and the peripheral site, as well as a third site. Note that the para-nitrophenol at the active site is flipped relative to the expected orientation; the position from which the phosphate is cleaved is facing away from the catalytic serine. There are two phosphate groups; one is in the catalytic site covalently bound to the catalytic serine, and the other is bound peripherally.
4KJG and 3MK1The third hPLAP image (3MK2.pdb) has a dark purple backbone and has the uncompetitive inhibitor, L-Phe, bound in the active site pocket. How would this prevent the catalytic events from occurring?
4KJG and 3MK2