Introduction
CRISPR Cas9 is an RNA-guided endonuclease that makes a sequence-specific double-stranded cut in DNA. Biochemistry experiments have revealed that the CRISPR Cas9 endonuclease
- Binds to the NGG PAM site on the Non-Target DNA strand
- Separates the Target- and Non-Target strands
- Interrogates the target strand with the spacer sequence at the 3’ end of its RNA
- If the Target DNA sequence matches the spacer sequence, two distinct nuclease active sites cut both the Target-DNA strand and the Non-Target DNA – strand 3 nucleotides upstream from the PAM site – resulting in a blunt-ended, double-stranded cut
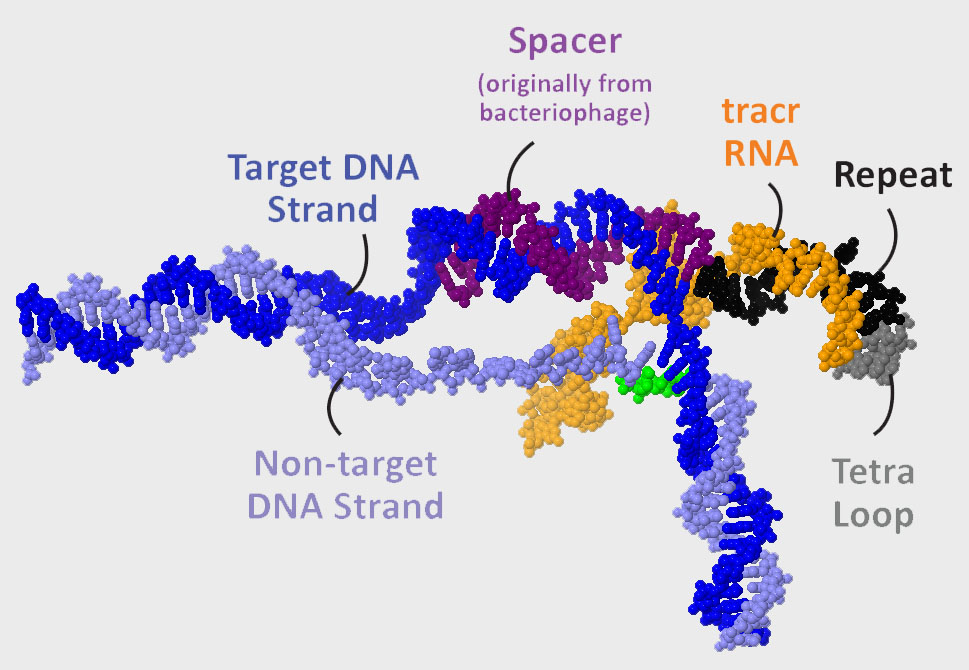
The guide RNA shown to the right is composed of tracr RNA (orange) and crRNA (crispr RNA). The 3’ end of the tracr RNA is complementary to the repeat sequence (black) found at the 5’end of the crRNA. Jennifer Doudna and colleagues converted this dual-guide RNA into a single-guide RNA by joining the 3’end of the tracr RNA to the 5’ end of the crRNA with a 4-nucleotide sequence known as the tetraloop (gray).
The spacer sequence that interrogates the Target-DNA strand extends from the repeat sequence to the 5’end of the single-guide RNA.
The Cas9 endonuclease is a large, conformationally dynamic protein that provides both the scaffold upon which this DNA/RNA complex forms and the two nuclease active sites that cut the DNA.
What are the protein domains of Cas9 that allows it to find and cut a DNA sequence that matches the spacer sequence of the guide RNA?

Cas9 Domains

The Cas9 protein is composed of 2 major domains: the Helical Domain and the Nuclease Domain. Together these two domains create two cavities large enough to accommodate the guide RNA, the Target DNA strand and the Non-target DNA strand.
Rotate the fully interactive 3D image to the right to explore this assembly.

The nuclease domain can be further broken down into the C-terminal domain , or CTD for short (light red), and RUVC domain (yellow).


As Cas9 searches for a viral sequence, it first binds to double-stranded DNA at a PAM site (NGG). This is accomplished by the PAM binding site located in a part of the CTD that is homologous to a topoisomerase II protein.


Upon PAM binding, Cas9 separates the two DNA stands and then uses the 3’end of its guide RNA to interrogate the Target-DNA strand


If the sequence of the guide RNA is complementary to the Target-DNA sequence, Cas9 undergoes a conformational shift in its 3D structure such that two different nuclease sites are activated. The Non-Target DNA strand is cut by the RUVC active site (red) located in the RUVC domain and the Target-DNA strand is cut by the HNH active site (green) located in the HNH domain.


Both DNA strands are cut 3 nucleotides away from the PAM site


But note that in the structure used in this Jmol Exploration (as well as for the physical model of Cas9) the HNH active site is far away from the cut site on the Target-DNA strand.


How do we resolve this apparent discrepancy in our story? The structure used here (based on 4un3.pdb) captured the Cas9 in a conformation that has not yet shifted to activate the HNH active site. In a later structure that captured the Cas9/guideRNA/DNA complex primed for cleavage (5f9r.pdb) a conformational shift has occurred that brings the HNH active site near the cut site on the Target-DNA strand.
Is this great science or what?
