Click Above to Play the Video

The Cas9 protein is quite large, about 1,350 amino acids long. This long chain of amino acids folds into a complex 3-dimensional shape, shown to the right. Click and drag on the image to rotate it in 3D.


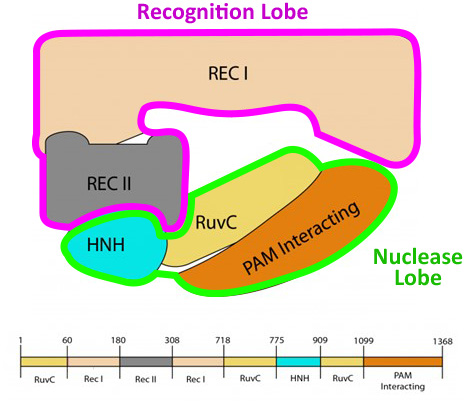
The protein is currently displayed in "backbone" format. But if we switch the display to "spacefill" format, in which every atom in the protein structure is shown as a sphere, we see two main regions: the recognition lobe, abbreviated REC, and the nuclease lobe, abbreviated NUC, each comprised of several smaller domains.


Individually, these domains are tightly folded globular shapes, with little space between them. But how tightly are they packed to each other? Do you see any gaps in between them?


There are indeed a number of small gaps between these five domains that make up the two main lobes. . . and as you might imagine, there are reasons for this. First, each domain is conformationally dynamic, meaning that they move independent of the other domains as the protein binds to target DNA and cuts it. Second, the cas9 protein binds to a folded RNA, called the “guide RNA”, shown in orange.


The sequence and 3D folded structure of the guide RNA has co-evolved with the Cas9 protein to fit tightly together. Anywhere it is not deeply embedded into the Cas9 protein, it folds back on itself to form hairpins stabilized by A-T/G-C base-pairs, highlighted in yellow.


But look closely. . . are there any areas of the guide RNA that are not deeply embedded into the Cas9 protein, and also not part of A-T/G-C base-pairs? Let’s temporarily remove the HNH domain of the Cas9 protein to make it easier to examine the entire guide RNA.


This one section here, about 20 RNA nucleotides long, looks relatively loose – with no A-T/G-C base-pairing and only barely kissing against the Cas9 protein in a couple spots.


Often when you find something odd or unique in a protein’s structure, it hints at what the protein might do, and how it does it. As it turns out, this exposed section of the guide RNA, called the “spacer”, is the magic behind the Cas9 protein. . . and we will discuss it in more detail in the next section.

Interrogating DNA
With the CRISPR Cas9 Protein
Click Above to Play the Video


To the right is the Cas9 protein (shown in white) tightly bound to the guide RNA comprised of a stable region (shown in orange) and a 20-nucleotide-long spacer region originally obtained from the genome of an attacking virus (shown in purple).


The Cas9 protein is expressed in the cytoplasm of the bacteria, where it is likely to encounter the genome of an invading virus. When Cas9 happens upon a section of DNA, it initially binds to it.


Once bound, if the Cas9 protein recognizes two guanine (G) nucleotides in a row, called the PAM sequence, it will uncoil the double stranded DNA. Is there anything you notice about either of these two separated strands of DNA?


One strand of the opened DNA, known as the “target strand”, lines up directly with the “spacer” of the guide RNA, almost as if the spacer is trying to test if it is complementary to the DNA, and can form A-T/G-C base-pairs with the target DNA strand.

Keeping in mind where the purple “spacer” section of the guide RNA originally came from, what would it mean if they were indeed complementary?


It would mean that the DNA being scanned matches the sequence of the virus that previously infected the bacteria, and therefore the same virus has come attacking again! Now what does that poor bacteria do?!

Making the Cut
With the CRISPR Cas9 Protein
Click Above to Play the Video


If the target DNA strand being compared to the purple spacer, then it is not from a previous virus, and that Cas9 simply lets go of it.


But, if it is complementary, the protein undergoes a change in shape that triggers the movement of the HNH and the RuvC domains close to the backbones of the two DNA strands.


Each of these two domains contains a separate nuclease active site that will cut a single strand of double stranded DNA exactly four nucleotides away from the NGG PAM Sequence.


This is a blunt ended double stranded cut, meaning there are no exposed unpaired nucleotides on either side of the cut site, making it incredibly hard for the DNA to be repaired, at least without introducing errors.


In this way, the amazing Cas9 protein serves as the judge, jury and executioner of this poor invading viral genome. As the judge, it lines up the evidence, positioning the DNA for examination. As the jury, it determines guilt, using the guide RNA to confirm that the DNA is indeed from an infecting virus. And as the executioner, it destroys the viral DNA, using two nuclease domains to create a blunt end cut in both strands of the DNA.

From Nature to BioTechnology
With the CRISPR Cas9 Protein
Click Above to Play the Video
