

What you see to the right is an antibody, with every single one of the over 10,000 atoms in this protein structure shown as a simple sphere, colored by element (carbon=gray, oxygen=red, nitrogen=blue, sulfur=yellow). Note that the antibody structure shown to the right is fully interactive and can be rotated in 3D by clicking and dragging the image.

While amazing in its complexity, this representation of an antibody is a bit visually overwhelming. So let's simplify it a bit. . . by coloring each of the four chains in this large protein a different color. Remember, some protein are comprised of only one chain of amino acids – but some, like antibodies, have multiple chains!
Color the antibody by chain.How many chains do you see? Do any of them look the same or do they all have a completely unique shape?
Antibodies are made up of four protein chains, two 'heavy chains' (shown in light and dark yellow) and two 'light chains' (shown in light and dark red). These four chains assemble together to form three distinct domains: the Fc domain, comprised of the bottom half of the two heavy chains, and two Fab domains, each comprised of one light chain and the top half of one heavy chain.
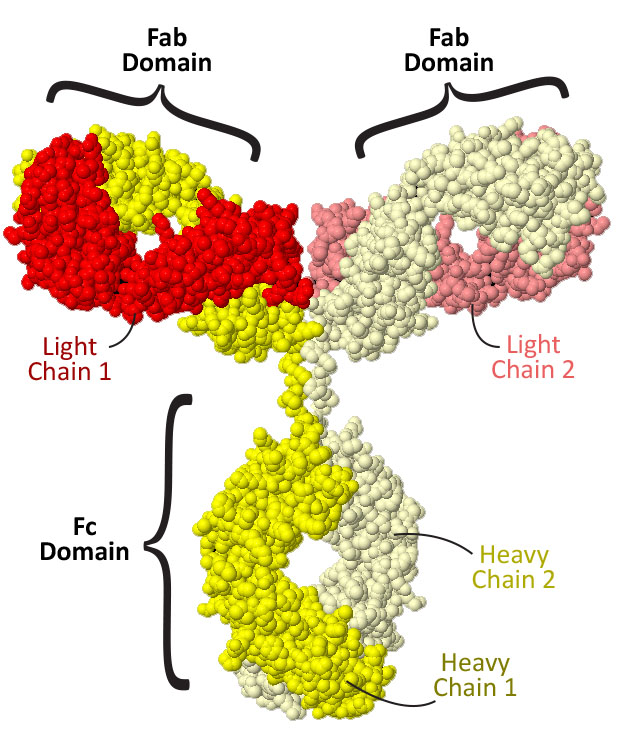
Do you think all three the domains are identical? Are any of them identical?
Even though we've simplified this representation by coloring it by chain instead of by individual atom element, it's still quite busy. So next let's remove all of the atoms except the single alpha carbon atoms at the center of each individual amino acid, and then connect these alpha carbons in order with a simple line.
Display the antibody as an alpha carbon backbone.This is called 'alpha carbon backbone' format, in that it highlights just the path of the backbone for each of the four folded protein chains of the antibody.
Now that the backbone is easier to see, do you notice any patterns? How does the backbone path of the two light chains compare? How about the two heavy chains? How about the heavy chains to the light chains?
Both the heavy chains and the light chains are made up of smaller motifs called 'IgG folds' that are linked together.
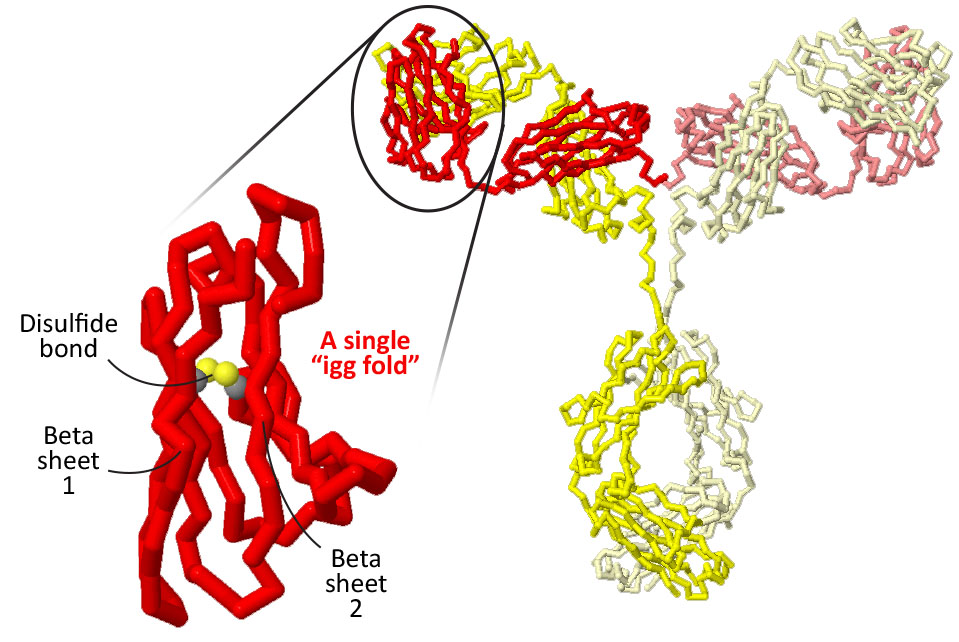
How many IgG folds are in each light chain? How many are in each heavy chain?
Each IgG fold is comprised of two beta pleated sheets held tightly together by a key disulfide bond that covalently connects two cysteine amino acids.
Focus in on a single 'IgG fold'.See if you can follow the path of the backbone from the beginning of the fold (highlighted in blue) to the end of the fold (highlighted in red).
Does the path of the backbone start with one full beta sheet and then progress to the second beta sheet? Or does the path of the backbone jump back and forth from one side to the other?
In addition to the key disulfide bonds found between the beta sheets of each of the twelve IgG folds, there are four additional disulfide bonds that help hold the four chains of an antibody together. Two are between the two heavy chains (near the hinge region), and two more are between each of the light chains and their corresponding heavy chains. Click the Jmol button below to display these four disulfide bonds on the antibody - and, using the schematic graphic below as a guide, see if you can find all four on the 3D structure.
Show the four additional disulfide bonds.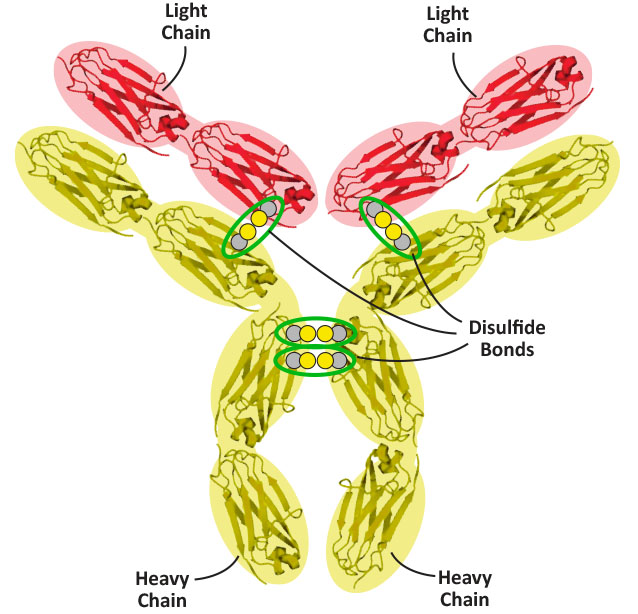
In the next activity, we will be exploring a single IgG fold in more detail, and creating a model that highlights the path of the backbone, the variable loops and the disulfide bond.
Save/Export Your Answers to the Questions in This Jmol Exploration